
Author: Arjun Suryawanshi
Mentor: Dr. Hong Pan
Unionville High School
Abstract
Metabolomics is a subset of bioinformatics and other -omics technologies which collects large swaths of data on biological samples. Specifically, metabolomics analyzes metabolites, small molecules present within the body as a result of metabolic pathways. Metabolites are valuable to analyze as they occupy the intersection between the environment and the body, demonstrating how important factors such as disease develop within people. Already, numerous diseases, from cancer to atherosclerosis, have shown the efficacy of metabolomic approaches as metabolites have been discovered which strongly correlate with the diseases, allowing for potential avenues of treatment. A core part of metabolomics is its multi-step methodology, including sample preparation, data collection, data processing, data analysis, and metabolite identification. A vast variety of mechanical and analytical approaches are necessary for all of these steps to occur. Of particular note is the role of machine learning in the data processing steps. Though common statistical methods are capable of processing, analyzing, and identifying metabolites from raw metabolomic data, artificial intelligence is capable of truly identifying patterns between the metabolites and accurately improving its own model. Nevertheless, there is still room for improvement in this area as many machine learning models have not been rigorously tested on a wide variety of datasets, limiting their applicability to metabolomics more broadly. Overall, metabolomics has been and will continue to be a valuable approach for making novel discoveries in medicine while advances in machine learning will continue to advance and enable the field.
Terms Table
Metabolic phenotyping – Characterizing a cell, organism, or biological system using metabolomics, describing the phenotype using chemical or metabolite readouts as a proxy for an organism’s observable biochemical traits
Endogenous metabolites – Metabolites/chemicals which are naturally synthesized within the body.
Exogenous metabolites – Metabolites/chemicals that are only naturally found in the environment and must enter through the body through environmental exposure, such as consuming food.
Machine Learning – A form of artificial intelligence that utilizes computer-based learning algorithms to find patterns or detect certain aspects of data. They broadly function by breaking down data into multidimensional vector points and then using error functions paired with layers of neurons that correspond to features to detect patterns or trends in the data and make conclusions.
Exposomics – A broader form of metabolomics that seeks to identify exogenous metabolites/chemicals in addition to endogenous ones.
Metabolome – All of the endogenous metabolites present within a certain region of the body or whole body. It’s akin to the genome encompassing all of the genes present within a cell/organism.
Exposome – Includes the metabolome as well as exogenous metabolites/chemicals that might be present.
Introduction
As you read this sentence, an uncountable number of chemical reactions are happening in your body. To move your eyes, to process these words, to digest your last meal, your body activates metabolic pathways, resulting in a vast number of molecules being synthesized or decomposed. These molecules are known as metabolites, and beyond being necessary for your body to function, they can also provide critical insights into your health. Metabolomics involves studying vast quantities of metabolites in order to create a dynamic picture of them in your body.
But why is simply knowing what molecules are in our body helpful? By collecting this data, metabolomics researchers can investigate the effects of the environment, disease, and medicine on our metabolites and extract medicinally relevant information. For example, knowing that a certain disease causes a unique molecule to increase may help doctors diagnose patients for that disease more quickly. A core aspect of metabolomics is artificial intelligence and machine learning, making the field a subcategory of the broader area of bioinformatics. As a computationally intensive and complex field of research, metabolomics has a critical intersection with modern data science approaches. Thus, in this review, metabolomics and its vast number of medicinal applications are explored and explained. Then, the value of artificial intelligence to metabolomics is discussed along with important future areas of research.
What is metabolomics
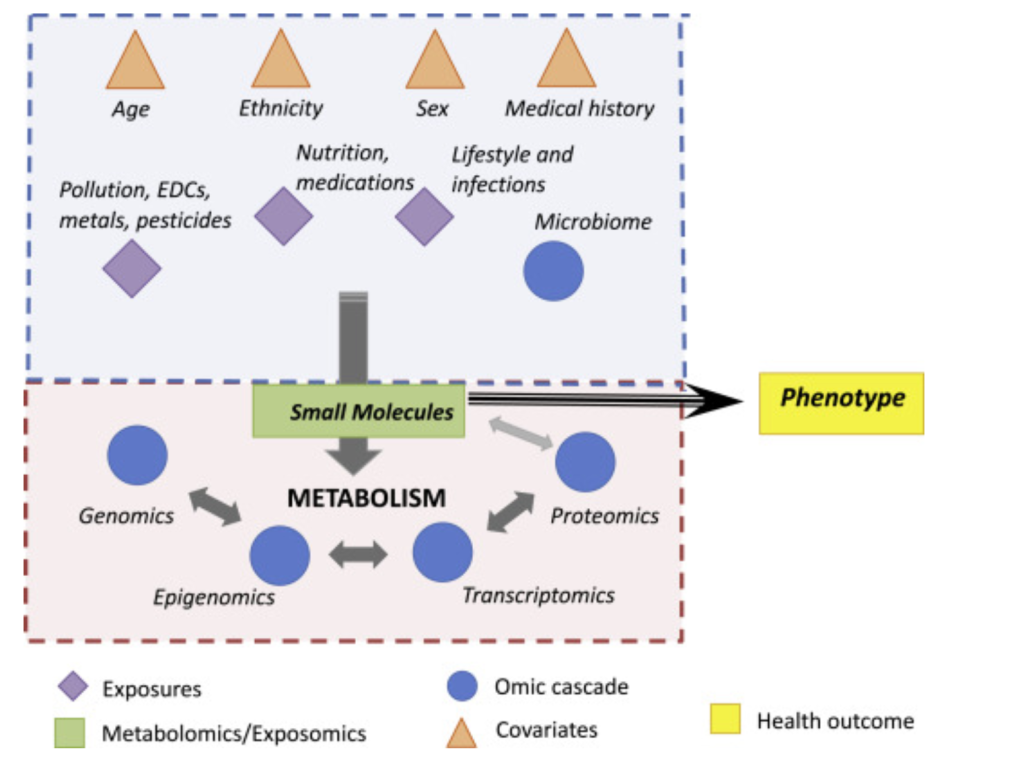
Metabolomics, like other -omics technologies such as genomics or proteomics, is a method of collecting and analyzing vast amounts of biological data (1). In this case, the molecule class of interest are metabolites, the products of reactants of metabolic pathways in a living organism.
For example, when glucose is broken down into pyruvate during glycolysis, the original molecule is converted into a number of other intermediate forms through each of the steps in glycolysis. Each of these intermediate molecules, in addition to glucose and pyruvate, are considered metabolites as they are the parts of a metabolic pathway, in this case glycolysis. Though proteins such as enzymes are parts of these pathways, they are excluded from metabolomics due to their size and are instead considered in proteomics.
Nevertheless, metabolomics is very broad in scope, encompassing more than 19,000 small molecules present in the human body due to enzymatic activity, food, medicine, the microbiome, and the environment (1). Before metabolomics became popular in research facilities, individual metabolites had been identified and used for disease identification. For example, diabetes is often detected using glucose test strips while levels of phenylalanine, an amino acid, in babies has been used to test for phenylketonuria (1). Now, with modern research techniques, hundreds or thousands of metabolites can be collected and analyzed from a small sample, leading to the rise of metabolomics.
The value of metabolites
As demonstrated in figure 1, metabolites are at the intersection of the environment, genotypic expression, and phenotype. Essentially, since the quantity and type of proteins present in the body is directly linked to the genome and since proteins affect the number of metabolites, metabolites serve as quantifiable products of gene expression (1). At the same time, the environment directly affects metabolites. Food, medicine, and pollutants can all introduce molecules into the body that affect metabolic pathways and the relative amount of metabolites, thus making metabolites simultaneously representative of environmental conditions. A good example of this metabolic interaction with the environment is the fact that 9/20 of the core amino acids humans use for protein synthesis come from food, tying together our metabolome with the environment.
Thus, to briefly summarize, metabolites are the downstream output of the genome and the upstream input from the environment, making metabolomics key to analyzing gene-environment interactions (3). This overlap is beneficial because other -omics technologies, particularly genomics, are unable to account for metabolites, preventing them from establishing the most direct link between genotype and phenotype (1). This is furthered by the fact that metabolites and their relative presence can be specific to tissues and change based on time while the genome is constant within each cell, thus making metabolomics an even more critical tool.
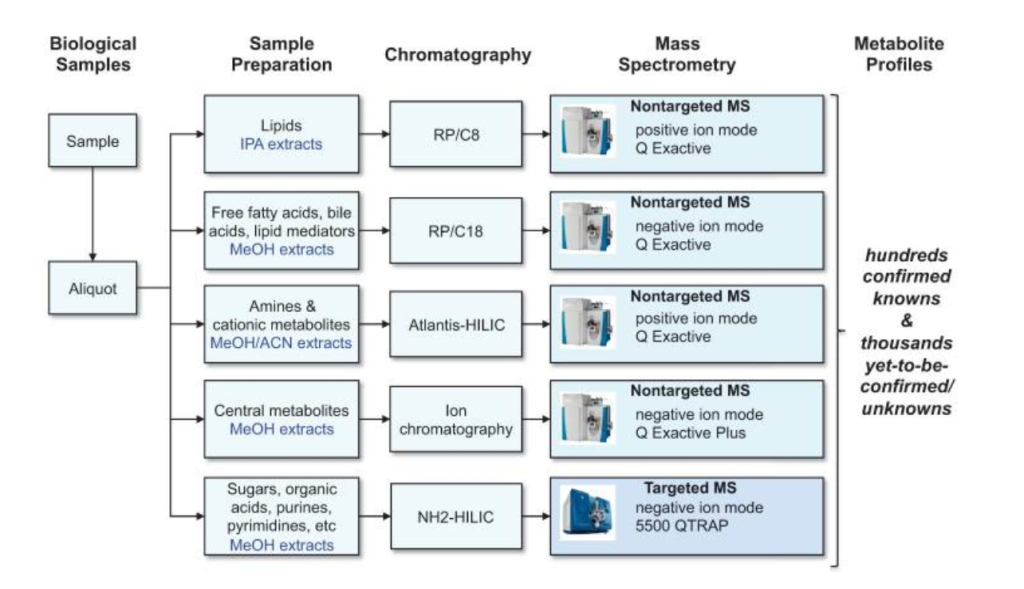
Metabolomic steps
As explained earlier, metabolomics is the metabolite version of an -omics approach, utilizing big data to collect metabolic information. However, it involves far more steps and instruments than other approaches due to the vast diversity of molecules from lipids to sugars (3). To summarize the process, a sample is first prepared from a biological sample such as a tissue. Then, a small aliquot or portion of it is taken and put through a form of chromatography, usually gas or liquid, in order to separate the molecules based on their size and retention time, enabling their discovery. Once they are separated by chromatography, they are put through a mass spectrometry machine that returns a mass to charge ratio to further pinpoint the exact molecule. After the entire sample goes through the mass spectrometry machine, relative abundance can also be discerned for all of the different molecules detected.
With all of this data, a number of analysis tools are then required to truly understand and pinpoint what molecules are actually in the sample and what might their relationship be to the broader experiment. Figure 2 summarizes this workflow for liquid chromatography and mass spectrometry (LC-MS). An alternative method of data collection is NMR spectroscopy which, though a fundamentally different methodology, produces spectra based on the molecules present and needs to be analyzed using a variety of tools (3).
Metabolomic subcategories
Though evidently complex, metabolomic methods hold great therapeutic potential (3, 7). In the past, many diseases were thought to have strong genetic components, making genomics a critical area of study. However, as more work has been done in the field, fewer mutations of interest have been identified that actually appeared to confer disease, resulting in reduced drug discovery. Instead, research is increasingly showing that environmental factors often significantly influence disease formation and progression, demonstrating the valuable role of metabolomics.
Another important point to note is that metabolomics has a wide variety of subcategories and names due to the vast number of methods and molecules to explore, from metabolic fingerprinting to metallomics (3). Of particular note for this review are exposomics, targeted metabolomics, and untargeted metabolomics. As discussed earlier, metabolites generally fall into two categories, ones produced within the body and those which enter from the environment, commonly referred to as endogenous and exogenous metabolites respectively. Exposomics focuses on both endogenous and exogenous metabolites as opposed to endogenous exclusively, as many metabolomics methods do (2). This broader focus allows for exposomics to be more encompassing and establish relationships between the environment and metabolic changes. However, many metabolites are still unknown to the exposome due to their low concentration or detectability, making a complete picture still unknown.
Targeted metabolomics focuses on a specific, predetermined set of metabolites across all samples, allowing for researchers to establish stronger relationships between experimental variables and metabolites affected (2). Untargeted metabolomics is the opposite of targeted, collecting data about all metabolites present. It is better at finding all potential metabolites or pathways affected though it increases the complexity of data analysis and provides less certainty than targeted metabolomics.
This is where Artificial Intelligence and Machine Learning can assist by pinpointing affected pathways and metabolites, potential biomarkers, or annotating unknown metabolites (2). As further elaborated later in this review, AI and ML can enhance multiple steps of metabolomic analysis. However, before exploring these areas, it is important to dive into greater specificity about the applications of metabolomics.
Where and how is/can metabolomics be applied?
As discussed previously, the usage of metabolites in medicine has had a fairly long usage, starting with fairly simple yet impactful techniques such as glucose test strips for diabetes (1). This simple example illustrates a critical aspect of metabolites, the fact that they show changes in the body as a result of some condition, be it eating food, exposure to a pollutant, or some genetic condition. Due to this, metabolomics has a variety of biomedical applications including determining the metabolic cause of a disease, medicine efficacy testing, novel drug targets, custom drug treatments, and biomarker discovery (1, 3, 4). This type of medicine is generally known as precision medicine as it can be both preventative and therapeutic while being tailored for an individual’s specific circumstances.
As demonstrated by numerous studies, metabolic changes are directly linked to a variety of disease mechanisms, pathogenic or otherwise (1). For example, metabolic changes have been proven to serve as early indicators for pancreatic cancer and type 2 diabetes. Furthermore, metabolic changes have demonstrated a connection between specific diets, such as those high in red-meats, and heart problems. Independently, metabolites can serve as biomarkers to test drug efficacy since medicines often interact with and affect metabolic pathways during digestion. Thus, by looking at the abundance of certain metabolites before and after taking a drug, researchers can determine what effect that drug is having, if it is functioning as intended or not, how quickly it is functioning, and a variety of other important metrics.
Atherosclerosis
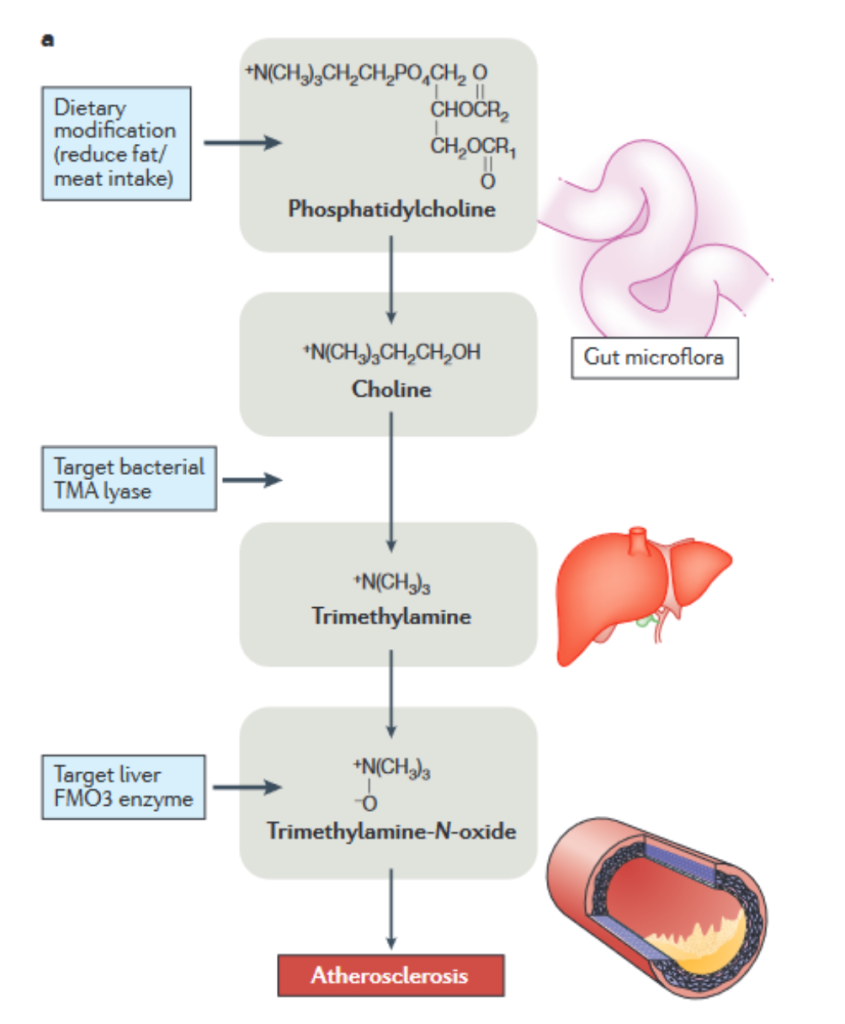
As mentioned earlier, previous approaches to drug discovery which focused on identifying target genes or proteins have been less successful due to many diseases not having strong genetic or inherited bases (3). On the other hand, metabolomics is more effective as it pinpoints much of the change that actually results in disease symptoms showing. For example, chronic diseases from Alzheimer’s disease to cancer have both been found to be correlated with significantly altered or dysregulated metabolism, providing an avenue for therapy.
Figure 3 visually demonstrates a more specific example of a metabolite that has been found to have a significant impact on disease development. Atherosclerosis, the development of fatty acid plaques within blood vessels, was previously thought to have a strong genetic component which was amplified by poor diet (3). However, an untargeted metabolomics study discovered a compound, Trimethylamine N-oxide (TMAO) that appeared to be strongly associated with atherosclerosis (3). This was further proven by the fact that rats injected with TMAO built up arterial plaques. Additionally, the metabolomic study was able to elucidate the pathway by which TMAO was formed. TMAO is produced from Trimethylamine (TMA) in the liver. TMA is formed by the breaking down of lipids such as cholines by gut microbiota. These lipids are found primarily in food, particularly meat. Additionally, by discovering this more complete metabolic picture of atherosclerosis development, researchers also discovered therapeutic compounds, such as 3,3-dimethylbutanol (commonly found in olive oil), which prevent TMAO synthesis.
Cancer
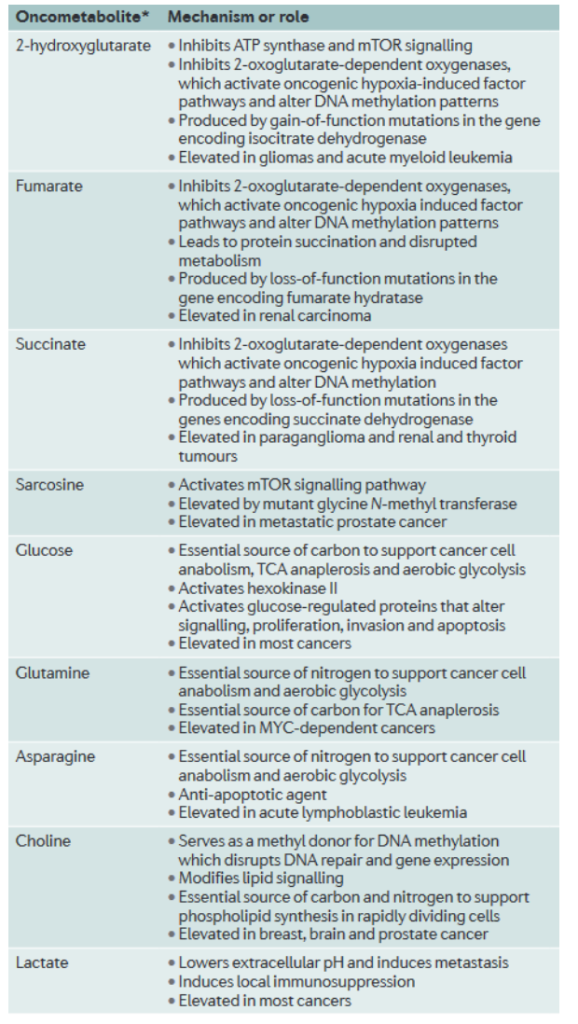
Metabolomics has also proven promising as a method for characterizing and treating cancers (3). Cancer is known to have a wide variety of causes, and though there are certain commonalities between cancers, such as certain oncogenes being mutated, they are often not the root cause and can’t solely be targeted for treatment. However, metabolomic studies have discovered that nearly all types of cancer fall into distinct ‘metabolic phenotypes’. Essentially, they have seven main mechanisms of metabolism and energy generation, including aerobic glycolysis, glutaminolysis, or one-carbon metabolism. By characterizing what type a cancer is, treatment can be customized to inhibit cancer development without harming somatic cells.
Furthermore, similar to oncogenes, oncometabolites have been identified which strongly affect tumor growth and metastasis (3). For example, metabolomic studies discovered 2-hydroxyglutarate to be elevated in gliomas (3). Once the metabolite was further analyzed, as shown in figure 4, it was discovered to inhibit ATP Synthase, activate an oncogenic hypoxia induced factor, and change DNA methylation patterns. These discoveries affirm the idea that many cancers take strong effect as metabolic disorders. Furthermore, other oncometabolites, also shown in figure 4, are not only associated with general metabolism, but specifically with cellular energy production. Fumarate, succinate, glucose, and lactate are all metabolites involved in cellular respiration, or the common method by which human cells produce energy. The fact that changes in the abundance of these metabolites then results in broader changes which enable cancer growth further suggests that analyzing and focusing on metabolic level treatments may be a more viable and effective approach to treat cancer.
How does AI/ML enhance metabolomics?
Now that metabolomics and its applications have been explored in sufficient depth, this section explores how artificial intelligence and machine learning can enable metabolomic studies. At its core, biology has a lot of variability and unpredictability due to the sheer volume of factors which may affect results. Machine learning approaches, such as neural networks, are excellent at dealing with unpredictability and big data as it can detect patterns by using hundreds if not thousands of layers of potentially relevant features (5).
Metabolomic Procedure
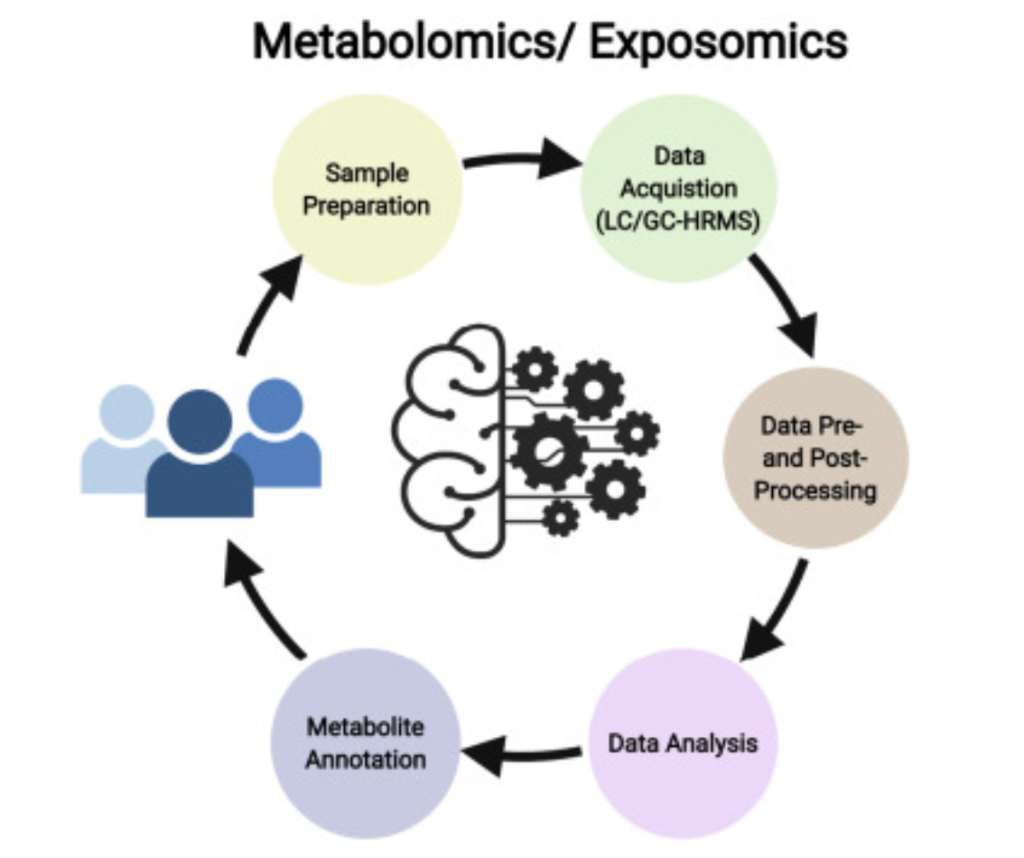
To explain all of the potential areas of application these approaches can have, it’s valuable to briefly elaborate on the procedure of metabolomics and clarify their necessary steps. As shown
in figure 5, the five core steps are sample preparation, data collection, data pre and post-processing, data analysis, and metabolite identification/annotation (2, 6). For sample preparation, solvents first separate proteins and other unwanted molecules from the rest of the metabolites. Then, for data collection, liquid or gas chromatography is performed in conjunction with mass spectrometry as discussed in previous sections. This step gives three key metrics, chromatography tells retention time while mass spectrometry tells mass-to-charge ratio and abundance. These first two steps are primarily enhanced by newer and improved instrumentation.
However, the next three are primarily computational tasks, allowing artificial intelligence to aid in the task (2). Data processing involves cleaning up, aligning, and organizing the data such that it’s able to be analyzed. Though LC and GC-MS are fairly effective, they do often have a lot of noise and molecule level issues that need to be resolved through processing. Data analysis primarily involves determining which spectra or signatures are relevant or actually demonstrate a significant change/difference from what was expected. Lastly, metabolite annotation and identification determines what metabolites/molecules the relevant features actually are, moving from a general class of molecule to a precise chemical formula and structure, allowing researchers to make precise conclusions based on the conditions of their experiment.
Data Processing
Data processing for metabolomics is often difficult due to the volume of data, number of potential interactions between metabolites, and somewhat noisy nature of mass spectrometry data (2). During pre-processing, researchers must first convert three measurements (retention time, mass-to-charge ratio, and abundance) into a two-dimensional system by combining retention time and mass-to-charge ratio. Then, by analyzing the area under the resulting peaks and combining with sample information, concentration of the molecule can be discovered. There are many softwares that currently do this job, but many of them have high false positive rates and don’t accurately combine peaks.
However, newer algorithms integrating machine learning approaches have demonstrated much higher accuracy though with some drawbacks (2). For example, a program known as WiPP developed in 2019 utilizes support vector machines, a type of machine learning model, to better classify higher concentration peaks though it is less effective for lower concentration metabolites. Additionally, programs using deep learning-based neural networks, such as PeakOnly or NeatMS have true detection rates in the high 90% though they slightly limit the range of concentrations they can work with. An even more effective model known as NPFimg actually converts gas chromatography-mass spectrometry data into 2-d image representations and then puts them through neural networks geared towards pattern recognition in images, resulting in true detection rates of more than 97%.
Data Analysis
The next step in the regular metabolomic workflow is data analysis, the process of determining which metabolites/chemicals are significantly correlated with the independent variable being tested (2). For example, figuring out which metabolites changed significantly in response to a drug intended to only affect a single metabolic pathway. Usually, univariate or multivariate linear statistical models are utilized to make these distinctions. They are capable of reaching conclusions, however, since their linear nature makes them unable to understand what the relationships between the metabolites are, causing higher rates of false positives and negatives.
Artificial intelligence overcomes this issue with its capacity to test different models and relationships between variables to understand what the most probable situation is, enabling identification of relationships between phenotypes and metabolites or biomarkers (2). A large variety of machine learning models have already been deployed for this task, ranging from random forest models to convolutional neural network architectures. They’ve also found success, from identifying metabolites associated with COVID-19 severity to lipids associated with Alzheimer’s disease progression. However, these approaches have primarily been applied to endogenous metabolites and not for exogenous ones, limiting their application to exposomics for the time being.
Metabolite Identification
Lastly, machine learning is valuable in determining what metabolite each peak precisely represents, allowing scientists to determine if they are reasonable and actionable discoveries (2). Currently, a large number of metabolite databases and spectral libraries exist and the exact results that a researcher gets may depend on the databases they choose to use since different laboratories and research organizations have different standards and primary libraries.
Most artificial intelligence approaches in this area seek to maximize the accuracy of a match between peak and metabolite by using additional factors of that molecule (2). In particular, by considering molecular structure, programs such as MetFrag and MIDAS predict resulting spectra for that molecule, allowing for comparison. Alternatively, programs such as Spec2Vec use this structural information to calculate the accuracy of a match between a metabolite and peak, resulting in higher accuracy than conventional methods. As with its application in other metabolomic steps, machine learning has not been applied much for unidentified and exogenous chemicals. Nevertheless, such models, particularly deep learning ones, will likely enable discovery in this area by combining data from existing databases and cross-applying that information to new metabolomic data.
What future areas of exploration are there for machine learning in metabolomics?
As suggested in the previous section, one of the primary areas of future research for artificial intelligence in metabolomics involves applying it to exogenous chemicals by testing such models on exposomic-based datasets (2). These datasets must also be far more diverse than
currently tested, enabling the model to improve and perform better for a wider variety of concentrations and samples. Additionally, expanding machine learning models such that they could even better predict spectral signatures exclusively based on chemical formula/structure could help characterize unknown chemicals. By improving metabolomics through artificial intelligence, the already proven medical value of metabolomics can be further actualized, enabling better understanding of disease, discovery of biomarkers, and creation of novel medicines (2,3).
Conclusion
To summarize, metabolomics is a big data -omics approach, most often using chromatography and mass spectrometry to collect massive amounts of data about the metabolites present in a sample. Metabolites are critical to a thorough understanding of disease as they exist at the intersection of the body and the environment, providing far more information about the body than knowledge about the genome or environment individually might give. As already proven for a variety of diseases and scenarios, metabolomics is a promising field for medicine as it not only discovers molecules relevant to diseases, but it also elucidates pathways associated with such molecules. In doing so, scientists can determine potential drug targets or solutions, as demonstrated with TMAO and 3,3-dimethylbutanol. Another critical aspect of metabolomics is its involvement with artificial intelligence and machine learning. Multiple steps of metabolomics, specifically data processing, data analysis, and metabolite identification all have and will continue to benefit from computational advances in machine learning. As more discoveries are made in the area, more accurate, precise, and relevant discoveries will be made on a metabolic level, furthering the medicinal application of metabolomics and its value to society.
Works Cited/Bibliography
1. Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015 Oct;1(1):a000588. doi: 10.1101/mcs.a000588. PMID: 27148576; PMCID: PMC4850886.
2. Petrick LM, Shomron N. AI/ML-driven advances in untargeted metabolomics and exposomics for biomedical applications. Cell Rep Phys Sci. 2022 Jul 20;3(7):100978. doi: 10.1016/j.xcrp.2022.100978. PMID: 35936554; PMCID: PMC9354369.
3. Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016 Jul;15(7):473-84. doi: 10.1038/nrd.2016.32. Epub 2016 Mar 11. PMID: 26965202.
4. Zhang GF, Sadhukhan S, Tochtrop GP, Brunengraber H. Metabolomics, pathway regulation, and pathway discovery. J Biol Chem. 2011 Jul 8;286(27):23631-5. doi: 10.1074/jbc.R110.171405. Epub 2011 May 12. PMID: 21566142; PMCID: PMC3129142.
5. Xu, C., Jackson, S.A. Machine learning and complex biological data. Genome Biol 20, 76 (2019). https://doi.org/10.1186/s13059-019-1689-0
6. Chen Y, Li EM, Xu LY. Guide to Metabolomics Analysis: A Bioinformatics Workflow. Metabolites. 2022 Apr 15;12(4):357. doi: 10.3390/metabo12040357. PMID: 35448542; PMCID: PMC9032224.
7. Johnson CH, Gonzalez FJ. Challenges and opportunities of metabolomics. J Cell Physiol. 2012 Aug;227(8):2975-81. doi: 10.1002/jcp.24002. PMID: 22034100; PMCID: PMC6309313.
About the author

Arjun Suryawanshi
Arjun is a senior at Unionville High School interested in pursuing biomedical engineering. In addition to BME, Arjun enjoys wet-lab research, debate, robotics, and science fair projects. In his free time, Arjun likes to play video games and gaze at the sky.
